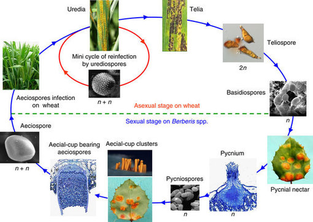
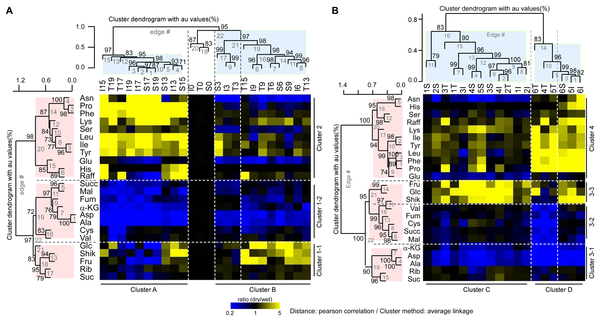
News and Updates
Reminder: Webinar Tomorrow on Pepper Breeding
If you haven't yet registered, there is still time to sign up for tomorrow's webinar, the latest in the NAPB webinar series. We would love for you to join us! The webinar is free but registration is required.
Start day/time: Tuesday, July 1, 2014 at 1:00 pm Eastern Time (-05:00 GMT)
Register: https://www1.gotomeeting.com/register/778576041
Title: Peppers are the Most Important Part of Salsa
About the Presenter
Dr. Kevin Crosby’s area of research at Texas A&M is plant breeding and genetics of vegetable crops. He has worked on melon, pepper, tomato, onion and carrot. The main emphasis of his research has been the elucidation of genetic mechanisms for stress tolerance and enhanced nutritional quality.
Reminder: Webinar Tomorrow on Dry Bean Breeding
If you haven't yet registered, there is still time to sign up for tomorrow's webinar, the latest in the NAPB webinar series. We would love for you to join us! The webinar is free but registration is required.
Start day/time: Tuesday, June 17, 2014 at 1:00 pm Eastern Time (-05:00 GMT)
Register: https://www1.gotomeeting.com/register/942418833
Title: Dry Bean Breeding isn't a Dry Topic
About the Presenter
Dr. James Kelly's dry bean breeding and genetics program is focused on the development of high yielding, disease and stress resistant cultivars with upright architecture, and improved canning quality in 10 commercial seed classes for production in Michigan.
2014 Plant Breeding Symposium: Global Food Security
Sponsored by DuPont Pioneer Hi-Bred and held at University of California Davis, the 2014 Plant Breeding Symposium is on Friday, April 11.
The event is also available as a live broadcast.
Please check the symposium website for more information about the line up of speakers and connecting to the webinar.
Upcoming Webinar - Identifying Soil Borne Diseases with Morphological and Molecular Techniques
Join us for a free webinar about morphological and molecular techniques for identifying soil born fungal and oomycete pathogens in dry bean production systems.
When: February 6, 2014 at 1:00pm Eastern Time (-05:00 GMT)
Presenter: Martin Chlivers, Michigan State University
Register now and learn more at http://www.extension.org/pages/68745.
Four Plant Genomic Conferences in Vienna this February
We are pleased to inform you that in February 2014, four conferences will be organized in Vienna by Vienna International Science Conferences & Events Association (VISCEA).
- Translational Cereal Genomics, Feb. 9-11, 2014;
- Plant Transformation Technology III, Feb. 12-14, 2014;
- Plant Gene Discovery & “Omics” Technology, Feb. 17-18, 2014;
- Applied Vegetable genomics, Feb. 19-20, 2014.
>>VISCEA is a non-commercial and non-profit organization, consisting of academy and industry scientists from across the world, founded in 2006 to initiate and assist in organizing professional and high quality scientific conferences and events. Vienna is one of the most beautiful cities in the world, situated in the heart of Europe. More information can be found at: www.viscea.org.<<
Thank you very much for your attention.
Svetlana Ujfalusi
Eucarpia Secretary
c/o Eidgenössisches Departement für Wirtschaft, Bildung und Forschung WBF
Forschungsanstalt Agroscope
Reckenholzstrasse 191
CH-8046 Zuerich
News: our new Website is online now. Go and have a look at www.eucarpia.org
Phone +41 44 377 71 88
Fax +41 44 377 72 01
E-mail to: eucarpia@art.admin.ch
Visit us: http://www.eucarpia.org
Plant Breeding for Drought Tolerance: Short Course June 2-13, 2014
Colorado State University will offer a three-credit, graduate-level, field-oriented course in Plant Breeding for Drought Tolerance, June 2-13, 2014. The course will be held in and around Fort Collins, Colorado, USA and is targeted to graduate students in the plant sciences, as well as to professionals in the public and private sectors interested in increasing their knowledge in this area.
Additional information on the course content, format, cost, and registration is available at http://www.droughtadaptation.org/summer_course.html. For questions, pleasecontact the Program Assistant, Kierra Jewell (Kierra.Jewell@ColoState.edu). Applications will be accepted through March 15, 2014 or until the class is full (20 students).
Breeding with Genomics Course at UC Davis February 11, 2014
The Seed Biotechnology Center at UC Davis is having their Breeding with Genomics Course on February 11 - 13, 2014.
PBG's own David Francis and our former content coordinator Shawn Yarnes are part of this event.
Below is some information provided by the Seed Biotechnology Center. Visit their webpage to learn more and to register! The early bird registration ends this Sunday, December 15.
Program
The course covers the basics of DNA markers, quantitative trait loci and the transition from maker assisted selection with a highlight on breeding for disease resistance.
New for 2014!
Extended modules on genomic selection, the latest integration of genomics to breeding, and a hands-on workshop on software support to marker application in breeding including the Integrated Breeding Platform and BLUPS.
Who should attend?
The course is aimed at professionals who are directly or indirectly involved in plant breeding and germplasm improvement. It is an opportunity for breeders who are already using these tools to expand their knowledge of new strategies and technologies and for laboratory personnel to appreciate how the marker data that they generate are applied in breeding programs.
Instructors
The course is taught by experts from both industry and academia. This is a great chance to interact with experts and technology specialists in plant breeding. They include:
Allen Van Deynze, UC Davis
Hamid Ashrafi, UC Davis
Kent Bradford, UC Davis
Jorge Dubcovsky, UC Davis
David Francis, The Ohio State University
Shawn Yarnes, The Integrated Breeding Platform Initiative
Graham McLaren, The Integrated Breeding Platform Initiative
Alison Van Eenennaam, UC Davis
December 2013
Having trouble viewing this? See it online.
Our Events
Publications of Interest
|
Reminder: Lattice Designs Webinar This Week
Join PBG for our webinar this week:
Title: Lattice Designs
When: Thursday, November 14 at 12:30 pm ET (New Time!)
Presenter: Dr. Jennifer Kling, Oregon State University
Registration Link: http://www.extension.org/pages/68608
Hope to see you there!
Find us on eXtension, Google+, Facebook, Twitter, or YouTube
November 2013
 News from Plant Breeding and Genomics on eXtension.org
News from Plant Breeding and Genomics on eXtension.org
|
From www.extension.org - Today, 9:01 AM
|
My time with the Plant Breeding and Genomics Community of Practice on eXtension.org has been a fantastic opportunity to see plant breeding from a multi-crop and multi-trait perspective. It has been a privilege to be involved in plant breeding extension through the development of open-source tutorials (http://www.extension.org/plant_breeding_genomics) and Plant Breeding and Genomics News (http://www.scoop.it/t/plant-breeding-and-genomics-news). I look forward to applying this experience in my new position as Genotyping Support Scientist with the Generation Challenge Program. Thank you to everyone who contributes to the Plant Breeding Community of Practice.
Sincerely, Shawn Yarnes |
Other Events
|
|
|
Recent Publications
|
From pubs.acs.org - October 15, 2:02 PM
|
|
|
From www.cell.com - October 23, 11:15 AM
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|